...
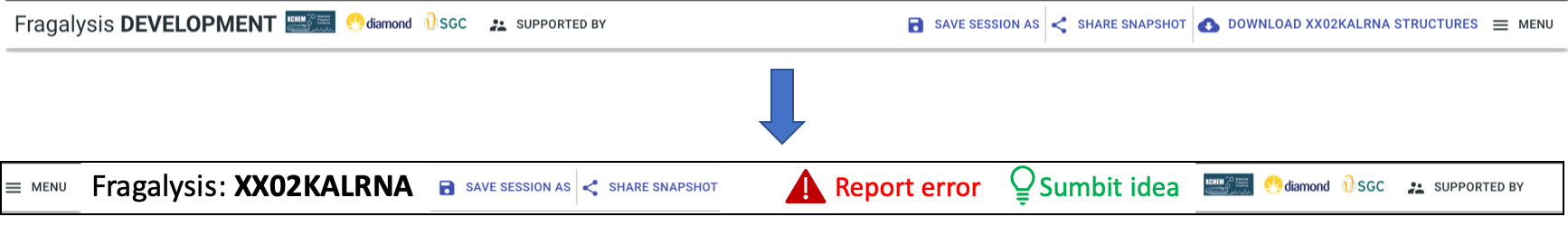
Header bar:Report: two prominent buttons, “Report error” (red), “Submit idea” (green)
Target name big and bold
Order along top: menu || Fragalysis: <target name> || session buttons (later “project”) || report buttons || logos etc.
Logos that are links should change the pointer (to a hand)
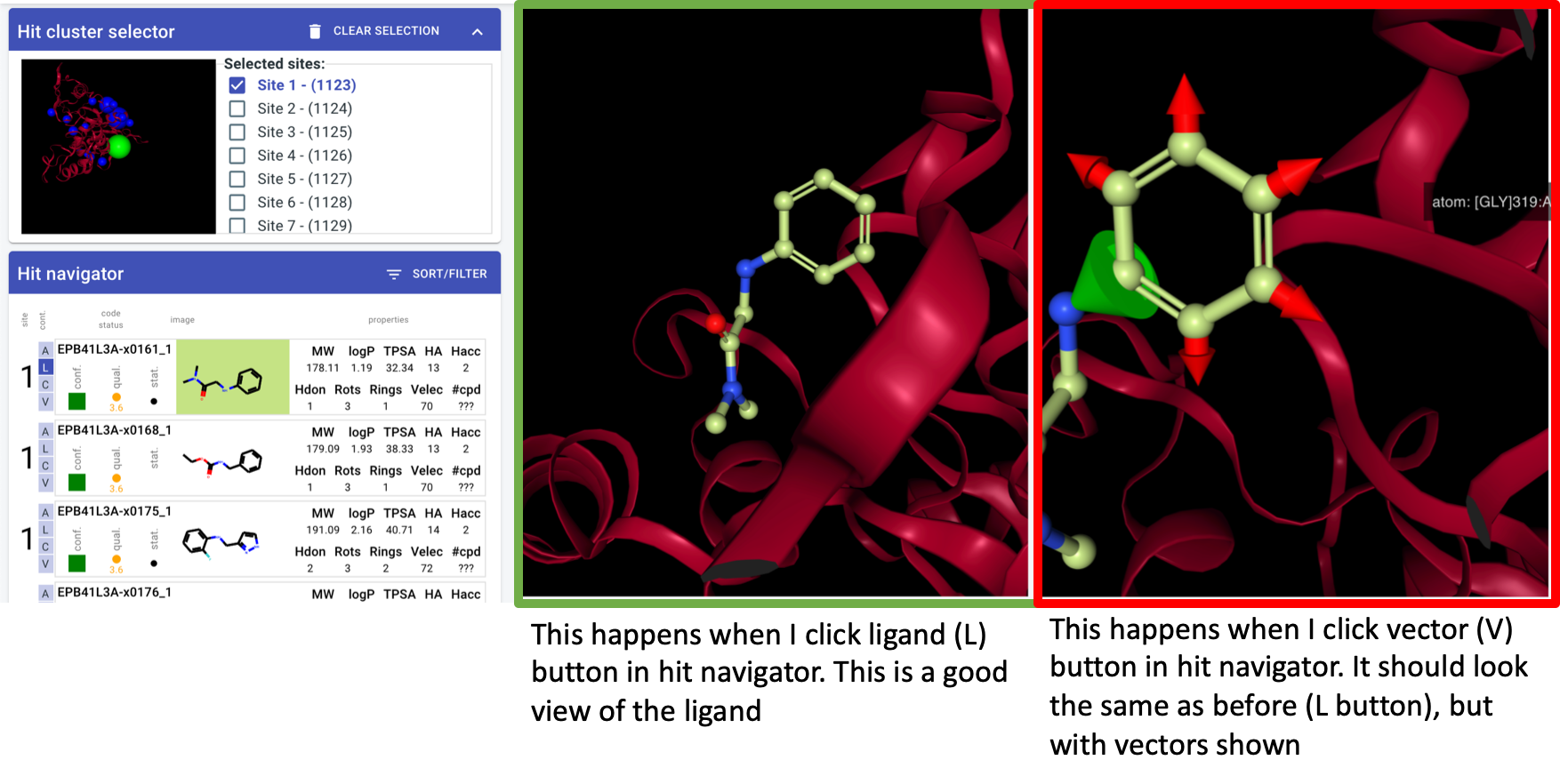
Site info in cluster navigatorDefaults on entry:
White background
Activate biggest cluster by default (most fragments)
Show all ligands in 3D viewer
Show complex of first ligand
(lazy loading of ligands?)
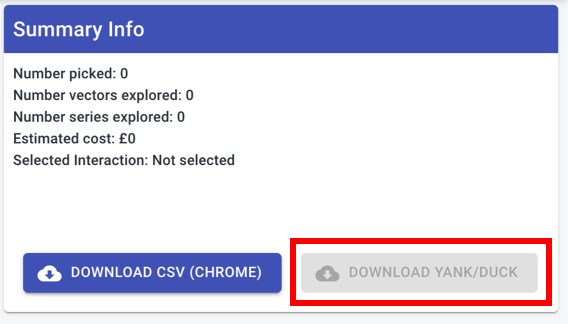
Remove download YANK button“
Clear selection” button: clear the filtersViewer controls:popup behaviour - like Office property editorget rid of header,3 buttons tight together
Automatic depth queuing - (pass them the backend code?)
orientation algorithm - v1: look down vector from centre of mass through centre of site
whatever the algorithm: should be consistent for a site regardless of which hit I click on
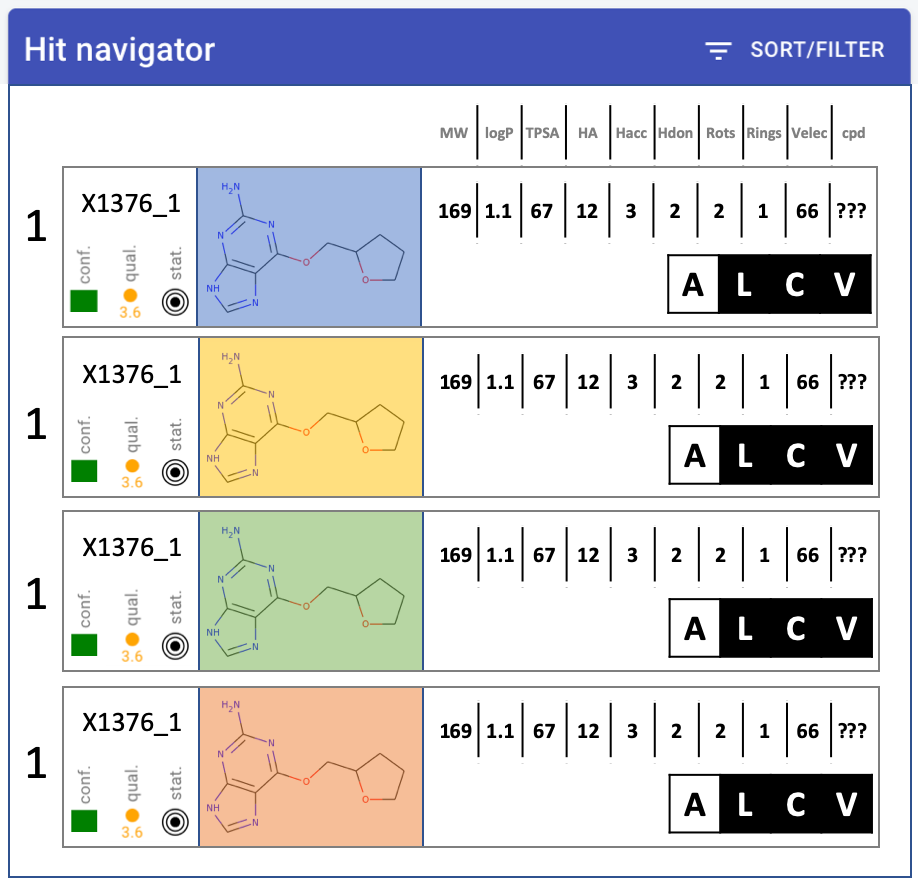
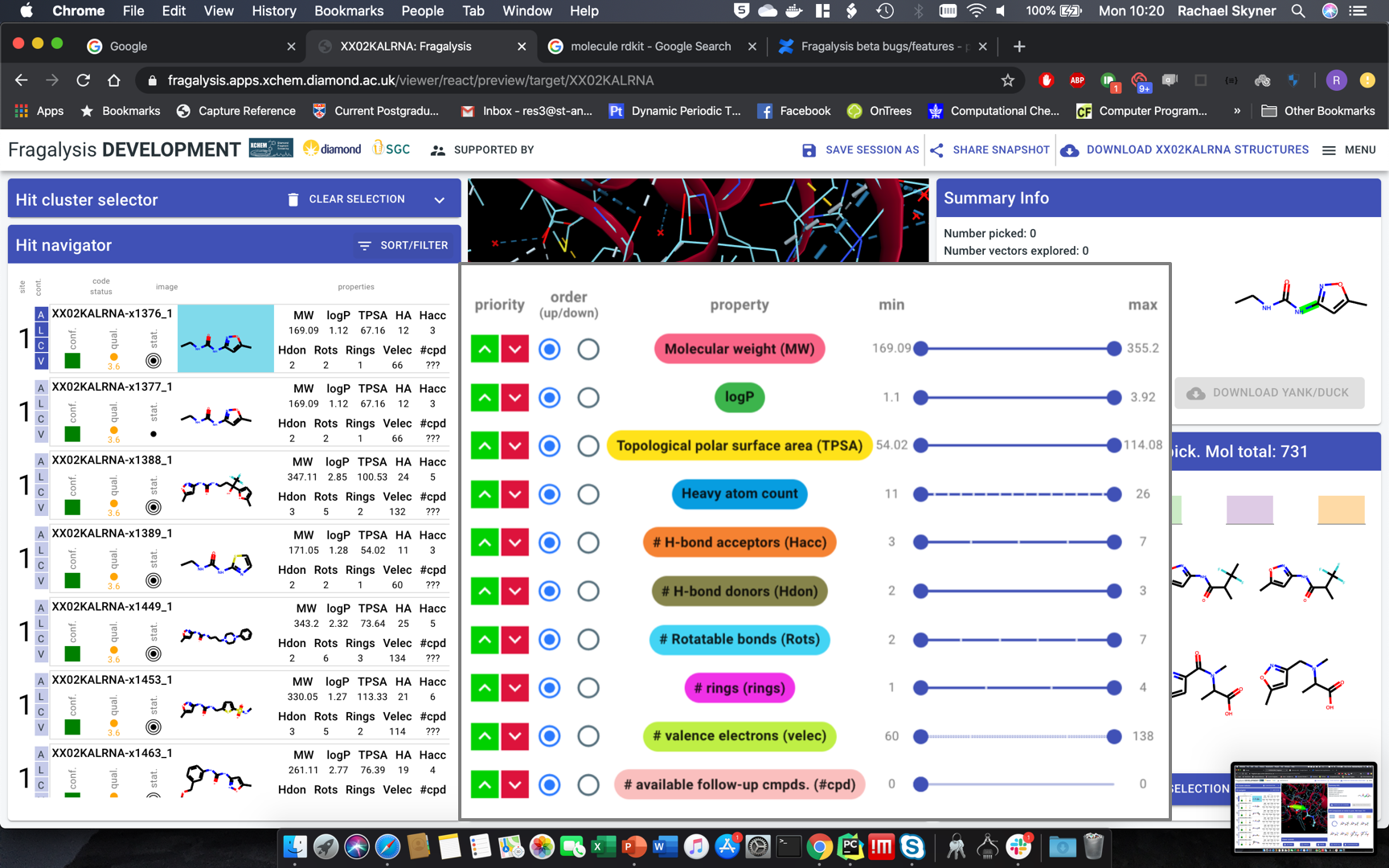
Hit navigator: layout(another mock-up)quality metrics - MUCH narrower, squish into single icon-letCompound much bigger, at least 2x widerChange properties into table: only one header at top, single rowRemove target name from labelNumbers - decimal points. none: MW, TPSA. one: logP#cpd - make it work - move to backend
#vectors - total (pickable)Velec - wrong number?
add electron density selector
display toggles: better contrast (check ICM, suggest you copy)
Hit navigator: drop-down to colour red thevalueoutside rule of 3 or 5 or none - left of sort/filter button. Default: rule-of-3.Sort-filter behaviour:slide out modal over 3D pane,withoutinactivating whole pageapply changes instantly, not on “apply”everything should remain active - just slide in-out.third the width (less than half): squish all icons, labels two rows if necessary, sliders far narrower
Hide all button in hit navigator - stay in site and 3D view
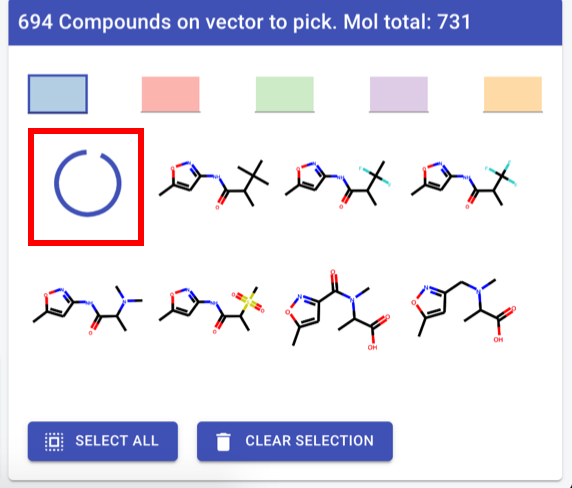
Compound picker
First compound in list: spinning wheelHighlight constant part - RDkit command
Header text: “<vector label>: <#cpds> available (total for molecule: <#total cpds>”Vector label: backend, for now human readable atom label straight from RDkit if that exists
if not: “vector 5” by whatever arbitrary sequence
ideally: can RDkit generate “alpha imine” or similar?
(the least amount of work possible)
Convert to table - exact same layout (and data) as hit navigator - including sort/filter
except quality things and label
sort/filter - slide out to left, but over 3D view)
display options: all, ligand, complex, vectors (hooks at least)
check-box on left - keep highlight of row if checked
by all means leave colours.
3D view - make it work
Header bar:Janssen logo
“supported by” - pop out a modal
(back-button from “supported by” resets the view - BUG. but eliminate through modal)
update logos in “supported by page”: ULTRA-DD, IMI, Horizon 2020,
...