Release (if possible)
Header bar:
Report: two prominent buttons, “Report error” (red), “Submit idea” (green)
Target name big and bold
Order along top: menu || Fragalysis: <target name> || session buttons (later “project”) || report buttons || logos etc.
Logos that are links should change the pointer (to a hand)
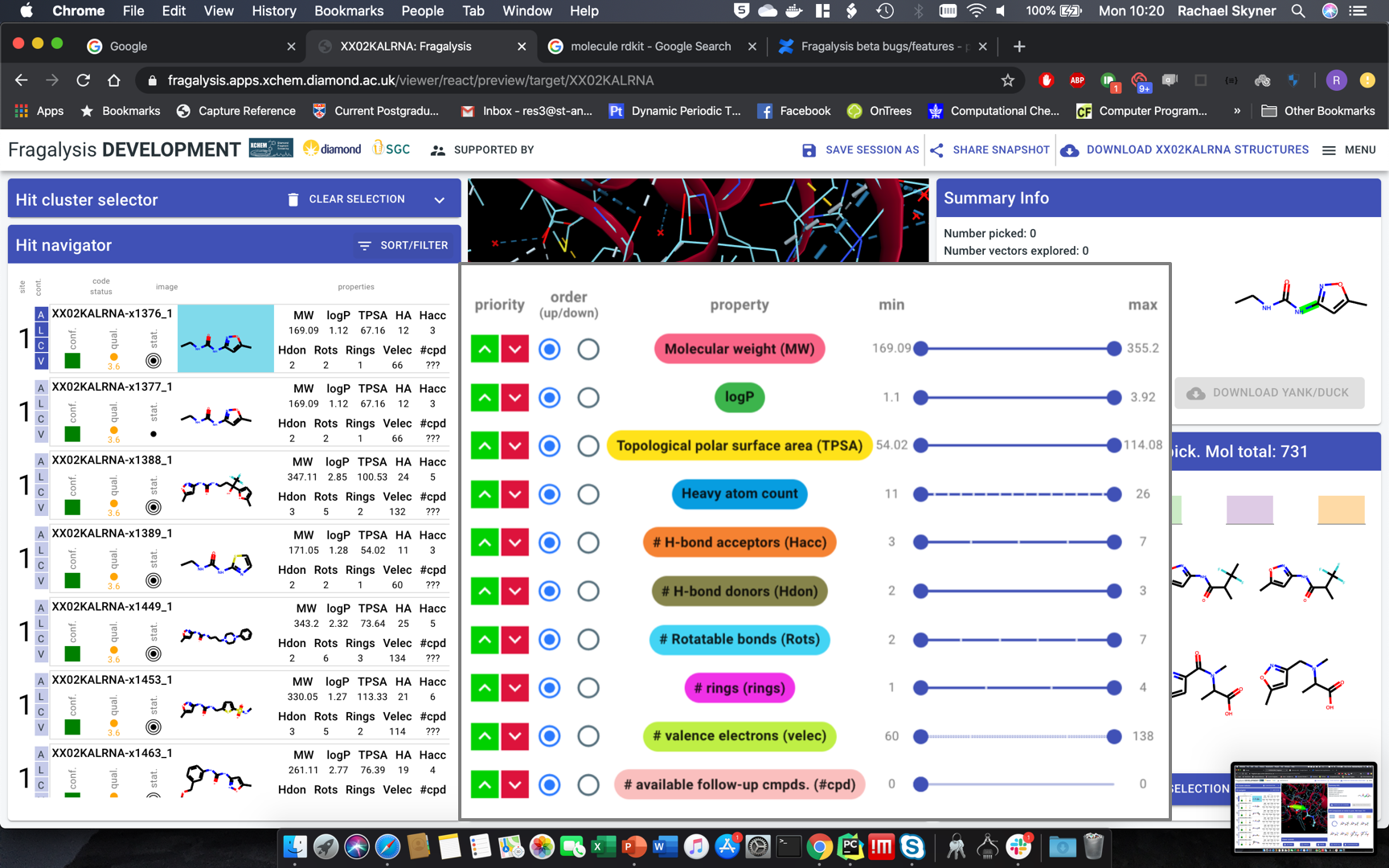
Site info in cluster navigator
Defaults on entry:
White background
Activate some cluster by default (first or biggest)
Activate first hit of first cluster, display all
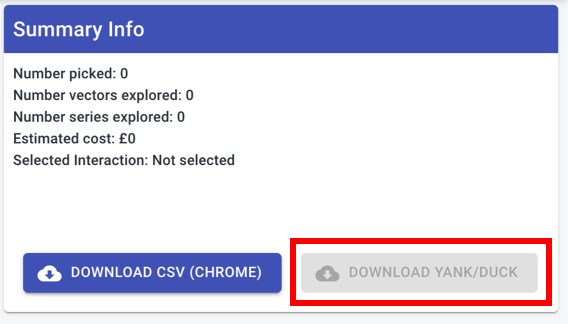
Remove download YANK button
“Clear selection” button: clear the filters
Viewer controls:
popup behaviour - like Office property editor
get rid of header,
3 buttons tight together
Automatic depth queuing - (pass them the backend code?)
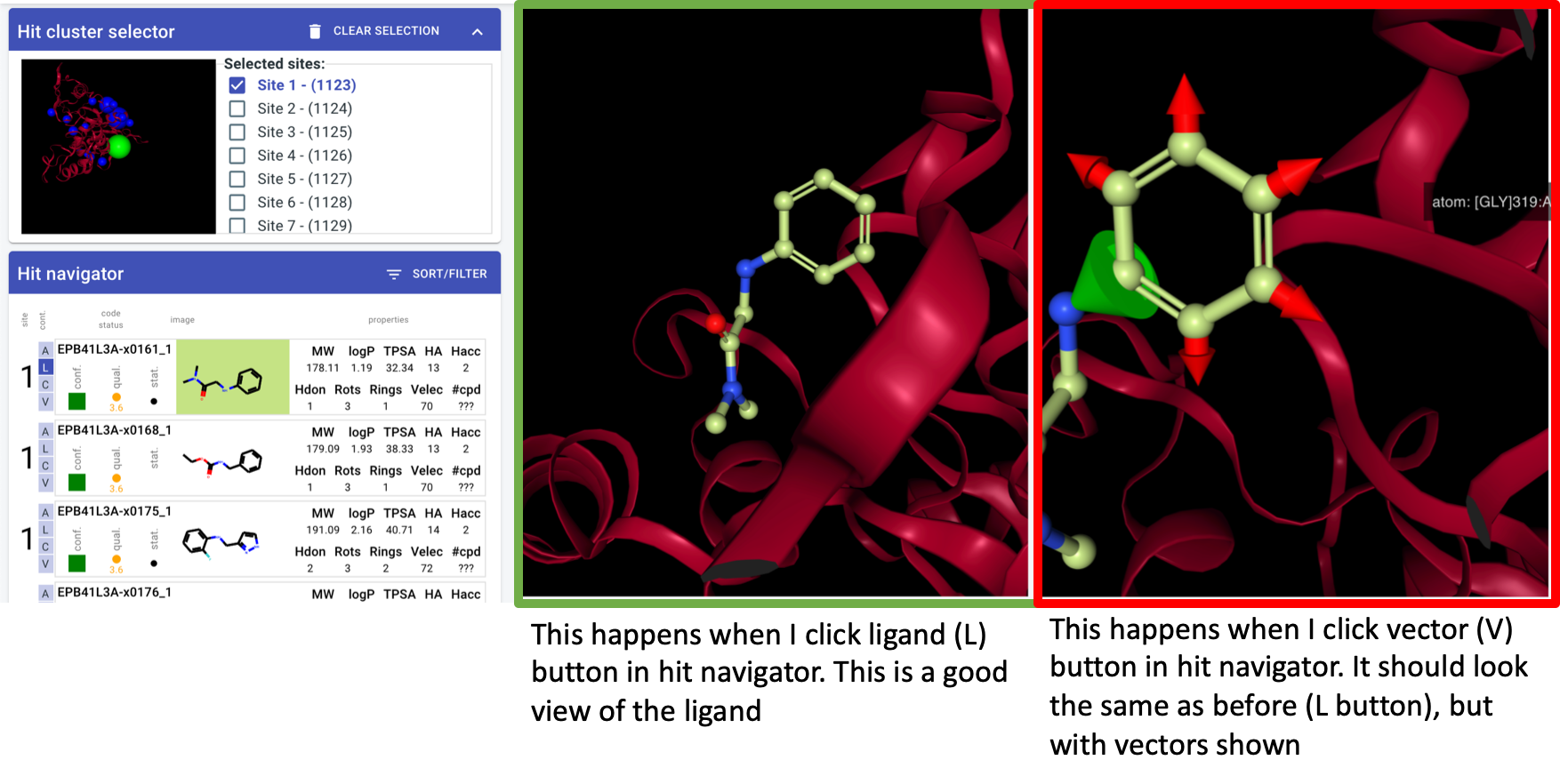
orientation algorithm - v1: look down vector from centre of mass through centre of site
whatever the algorithm: should be consistent for a site regardless of which hit I click on
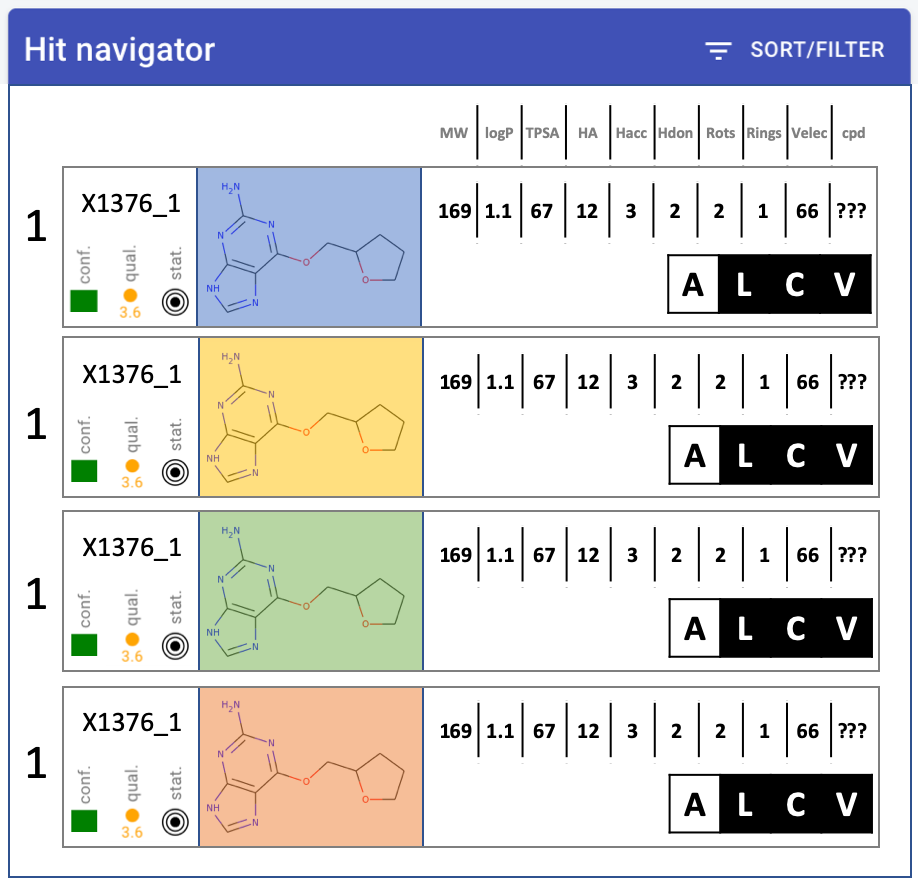
Hit navigator: layout
(another mock-up)
quality metrics - MUCH narrower, squish into single icon-let
Compound much bigger, at least 2x wider
Change properties into table: only one header at top, single row
Remove target name from label
Numbers - decimal points. none: MW, TPSA. one: logP
#cpd - make it work - move to backend
#vectors - total (pickable)
Velec - wrong number?
add electron density selector
display toggles: better contrast (check ICM, suggest you copy)
Hit navigator: drop-down to colour red the value outside rule of 3 or 5 or none - left of sort/filter button. Default: rule-of-3.
Sort-filter behaviour:
slide out modal over 3D pane, without inactivating whole page
apply changes instantly, not on “apply”
everything should remain active - just slide in-out.
third the width (less than half): squish all icons, labels two rows if necessary, sliders far narrower
Hide all button in hit navigator - stay in site and 3D view
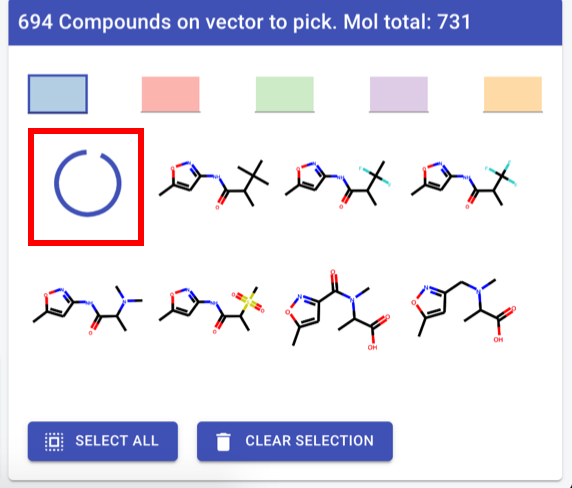
Compound picker
First compound in list: spinning wheel
Highlight constant part - RDkit command
Header text: “<vector label>: <#cpds> available (total for molecule: <#total cpds>”
Vector label: backend, for now human readable atom label straight from RDkit if that exists
if not: “vector 5” by whatever arbitrary sequence
ideally: can RDkit generate “alpha imine” or similar?
(the least amount of work possible)
Convert to table - exact same layout (and data) as hit navigator - including sort/filter
except quality things and label
sort/filter - slide out to left, but over 3D view)
display options: all, ligand, complex, vectors (hooks at least)
check-box on left - keep highlight of row if checked
by all means leave colours.
3D view - make it work
Header bar:
Janssen logo
“supported by” - pop out a modal
(back-button from “supported by” resets the view - BUG. but eliminate through modal)
update logos in “supported by page”: ULTRA-DD, IMI, Horizon 2020,
Wishlist
Landing page, Target List - more columns
target info (Uniprot, anything) - auto-generate starting from reference structure sequence
link to pdb group deposition page (if it is a thing - else something similar - might be query)
wiki page (if exists)
(Brian will have suggestions)
Display electron density - extra display selector
Colour compound properties - backend for implementing rules
future: customise on the fly (conditional formatting, or like sort/filter thingy)
move filters into backend
Compound navigator
Convert to tree-view to navigate sub-vectors
Download CSV button
Richer display of 2D compounds - map RDkit properties onto 2D view
Ligand interaction diagram - comparison tools. (Needs thinking)